Biophysics & Nanoscience
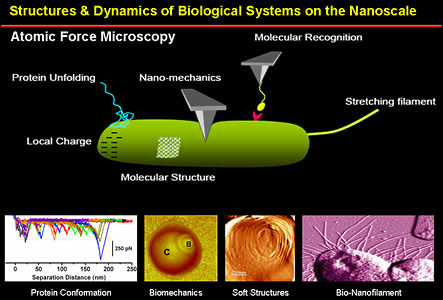 The Biophysics and Nanoscience group in our department is represented by Dr. Natalia Guevara, Dr. Juan Guevara, Dr. Andreas Hanke, Dr. HyeongJun Kim, and Dr. Ahmed Touhami. They are involved in several multidisciplinary projects including fundamental aspects of DNA conformational changes and protein binding, dynamics and spectroscopy of a single DNA molecule, nanostructural and nanomechanical properties of biological surfaces, manipulating individual molecules in living cells, and Low-density Lipoprotein (LDL)/DNA interactions. The goals are not only to establish a fundamental understanding of living processes at the molecular level but also to help in the development of new biologically-inspired materials. These highly multidisciplinary projects provide numerous research and training opportunities for undergraduate and graduate students to work at the interface of physics, chemistry, biology, and Nanoscience.
The Biophysics and Nanoscience group in our department is represented by Dr. Natalia Guevara, Dr. Juan Guevara, Dr. Andreas Hanke, Dr. HyeongJun Kim, and Dr. Ahmed Touhami. They are involved in several multidisciplinary projects including fundamental aspects of DNA conformational changes and protein binding, dynamics and spectroscopy of a single DNA molecule, nanostructural and nanomechanical properties of biological surfaces, manipulating individual molecules in living cells, and Low-density Lipoprotein (LDL)/DNA interactions. The goals are not only to establish a fundamental understanding of living processes at the molecular level but also to help in the development of new biologically-inspired materials. These highly multidisciplinary projects provide numerous research and training opportunities for undergraduate and graduate students to work at the interface of physics, chemistry, biology, and Nanoscience.
DNA and Proteins.
Areas of Research
- Low-density lipoprotein mediated cell transfection
 Elevated plasma levels of low-density lipoprotein, LDL, correlate with cardiovascular diseases such atherosclerosis, diabetes, and stroke. Our research is focused on the elucidation of the function of apo B100, the major apolipoprotein in LDL, VLDL, and other low-density lipoproteins. The primary structure of apo B100 was determined chemically by the laboratories of Drs. Chao-Yuh Yang and Lawrence Chan at Baylor College of Medicine in the mid 1980’s. Apo B100 is a massive protein of 4,536 amino acids with few clues as to functions other than binding lipids. We have identified regions in the apo B100 sequence, which are similar or analogous to important domains present in signaling proteins, transcriptional factors, ribonuclear proteins, and viral proteins. We shown experimentally that LDL binds nucleic acids and has the capacity to transport and deliver DNA in vitro using a variety of cell types and in vivo in the rat. We have evidence that LDL from individuals who were seropositive for the Human Cytomegalovirus (HCMV) carries the viral genome and other investigators have similar evidence obtained from individuals burdened with the hepatitis c virus. We have experimental evidence that two regions of the apo B100 molecule bind DNA and synthetic peptides of one region functions like the capsid proteins of the Flaviviridae viruses, hepatitis c, yellow fever, West Nile, and dengue viruses. Our observations strongly suggest that apo B100 is a member protein of the immune response. Our hypothesis is that lipids which have been shown to mediate the conformation of the apo B100 molecule are important in LDL’s role in removing nucleic acids from invading microbes from circulation.
Elevated plasma levels of low-density lipoprotein, LDL, correlate with cardiovascular diseases such atherosclerosis, diabetes, and stroke. Our research is focused on the elucidation of the function of apo B100, the major apolipoprotein in LDL, VLDL, and other low-density lipoproteins. The primary structure of apo B100 was determined chemically by the laboratories of Drs. Chao-Yuh Yang and Lawrence Chan at Baylor College of Medicine in the mid 1980’s. Apo B100 is a massive protein of 4,536 amino acids with few clues as to functions other than binding lipids. We have identified regions in the apo B100 sequence, which are similar or analogous to important domains present in signaling proteins, transcriptional factors, ribonuclear proteins, and viral proteins. We shown experimentally that LDL binds nucleic acids and has the capacity to transport and deliver DNA in vitro using a variety of cell types and in vivo in the rat. We have evidence that LDL from individuals who were seropositive for the Human Cytomegalovirus (HCMV) carries the viral genome and other investigators have similar evidence obtained from individuals burdened with the hepatitis c virus. We have experimental evidence that two regions of the apo B100 molecule bind DNA and synthetic peptides of one region functions like the capsid proteins of the Flaviviridae viruses, hepatitis c, yellow fever, West Nile, and dengue viruses. Our observations strongly suggest that apo B100 is a member protein of the immune response. Our hypothesis is that lipids which have been shown to mediate the conformation of the apo B100 molecule are important in LDL’s role in removing nucleic acids from invading microbes from circulation.
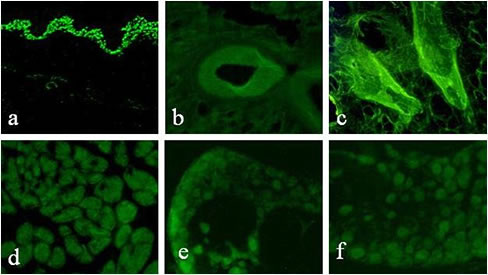
LDL-mediated GFP gene Delivery in vivo Frozen sections from transfected rats
Research Faculty
Single-molecule biophysics.
Recent advances in the detection and manipulation of single molecules offer great promise for investigating the structural distributions and dynamics of complex systems. We develop and use single-molecule methods to address key issues in protein, nucleic acid and cellular dynamics and function. These novel methods provide unprecedented abilities to directly probe static and dynamic heterogeneity in biological and cellular systems, under both equilibrium and non-equilibrium conditions. For example, directly observing multiple steps and pathways in protein folding and misfolding, real time visualizing and investigating the cell growth and division in single bacterium, and directly observing and testing mechanisms of antimicrobial peptides in live cells are some of the important capabilities of single-molecule methods that we are developing and applying in our research.
Our current research is focused on the following areas:
- Bacterial structures and dynamics
- Mechanics of biological filaments
- Protein folding, misfolding, and aggregation
- Lipid-Protein Interaction
- Statistical mechanics of biomolecules
- Protein-DNA interaction
Bacterial structures and dynamics.
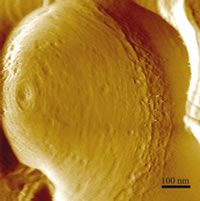 We are combining atomic force microscopy (AFM) with fluorescence methodologies to investigate the structural and physical properties of different microbial living cells in their physiological medium in order to explore at sub-nanometer resolution the cell-surface dynamics such as flexible protein domains and growth processes (peptidoglycan network, cell wall turnover, and division phenomena). Spatially and temporally synchronized atomic force and total internal reflection fluorescence microscopy is used to monitor time-dependent processes such as surface morphology changes related to the interaction between the cell wall compounds and external agents (enzymes, antibodies, and drugs).
We are combining atomic force microscopy (AFM) with fluorescence methodologies to investigate the structural and physical properties of different microbial living cells in their physiological medium in order to explore at sub-nanometer resolution the cell-surface dynamics such as flexible protein domains and growth processes (peptidoglycan network, cell wall turnover, and division phenomena). Spatially and temporally synchronized atomic force and total internal reflection fluorescence microscopy is used to monitor time-dependent processes such as surface morphology changes related to the interaction between the cell wall compounds and external agents (enzymes, antibodies, and drugs).
Mechanics of biological filaments.
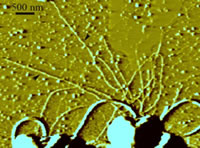 Pili are thin polymeric filaments that extend several micrometers from the bacteria cell envelope. They are involved in binding not only to host mammalian cells but also to abiotic surfaces; further, they are important for surface motility, biofilm formation, and horizontal gene transfer. We are using Optical Tweezers (OT) and Total Internal Reflection Fluorescence (TIRF) techniques to explore and observe how do these molecular machines move, bind, and unbind? How are their mechanical properties affected by changing physiological and environmental conditions? The future of these kinds of experiments lies in the detailed understanding of interfacial interactions, such as cell-support, cell–cell, and cell–drug interactions.
Pili are thin polymeric filaments that extend several micrometers from the bacteria cell envelope. They are involved in binding not only to host mammalian cells but also to abiotic surfaces; further, they are important for surface motility, biofilm formation, and horizontal gene transfer. We are using Optical Tweezers (OT) and Total Internal Reflection Fluorescence (TIRF) techniques to explore and observe how do these molecular machines move, bind, and unbind? How are their mechanical properties affected by changing physiological and environmental conditions? The future of these kinds of experiments lies in the detailed understanding of interfacial interactions, such as cell-support, cell–cell, and cell–drug interactions.
Protein folding, misfolding, and aggregation.
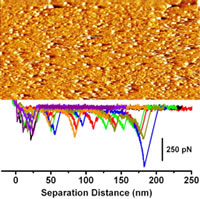 Proper folding is vital for proteins to function. A wrong fold changes the shape of the protein, which can alter how it interacts with other molecules. If the folding goes wrong, the protein can stop working, or worse, act in harmful ways. For example, Alzheimer's disease is caused by a misfolded protein. We are particularly interested in Light Chain Amyloidosis, a misfolding disease characterized by the deposition of monoclonal immunoglobulin light chains as amyloid fibrils which affect several organs, causing dysfunction.
Proper folding is vital for proteins to function. A wrong fold changes the shape of the protein, which can alter how it interacts with other molecules. If the folding goes wrong, the protein can stop working, or worse, act in harmful ways. For example, Alzheimer's disease is caused by a misfolded protein. We are particularly interested in Light Chain Amyloidosis, a misfolding disease characterized by the deposition of monoclonal immunoglobulin light chains as amyloid fibrils which affect several organs, causing dysfunction.
Lipid-Protein Interaction.
 MinD is a peripheral member ATPase involved in the selection of the division site in Escherichia coli. Its activity is stimulated by MinE but only in the presence of phospholipids and MinC. We are using vesicles of synthetic and natural phospholipids as a model system for these studies. AFM-based single molecule force spectroscopy combined with fluorescence technique allows us to investigate the different interactions between the Min system proteins and the phospholipids in real time and at single molecule resolution.
MinD is a peripheral member ATPase involved in the selection of the division site in Escherichia coli. Its activity is stimulated by MinE but only in the presence of phospholipids and MinC. We are using vesicles of synthetic and natural phospholipids as a model system for these studies. AFM-based single molecule force spectroscopy combined with fluorescence technique allows us to investigate the different interactions between the Min system proteins and the phospholipids in real time and at single molecule resolution.
Research Faculty
Casimir Effect.
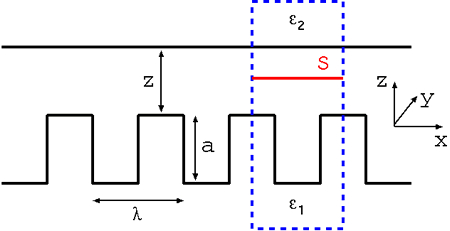 The increasing miniaturization of nanomechanical systems requires an understanding of fundamental forces on the nanoscale. When the distance between metallic or dielectric surfaces is reduced to the submicron range, forces of pure quantum electrodynamical origin emerge. This force was first predicted 1948 by H. B. G. Casimir for two infinitely extended, perfectly conducting, electrically neutral plates in vacuum. The Casimir force can be understood as resulting from the modification of the quantum fluctuations of the electromagnetic field in vacuum by the presence of boundaries. Due to its nature as a fluctuation-induced force, the Casimir force has a strong dependence on both the geometry and the optical properties of the interacting bodies.
The increasing miniaturization of nanomechanical systems requires an understanding of fundamental forces on the nanoscale. When the distance between metallic or dielectric surfaces is reduced to the submicron range, forces of pure quantum electrodynamical origin emerge. This force was first predicted 1948 by H. B. G. Casimir for two infinitely extended, perfectly conducting, electrically neutral plates in vacuum. The Casimir force can be understood as resulting from the modification of the quantum fluctuations of the electromagnetic field in vacuum by the presence of boundaries. Due to its nature as a fluctuation-induced force, the Casimir force has a strong dependence on both the geometry and the optical properties of the interacting bodies.
In 2001, researchers at Bell Labs pioneered experiments that demonstrated the possibility of using the Casimir force as an actuation force of micromechanical components. These experiments establish the importance of quantum electrodynamical effects in micro- and nanomechanical systems (MEMS and NEMS), hence the convergence of fundamental physics and real world technological applications. Concurrently with this development, significant progress has been made in the last decade in predicting the Casimir force for complex geometries and materials, as often encountered in actual nanomechanical systems.
Computational cell for the calculation of the electromagnetic Casimir force between
a flat surface and nanoscale trench arrays by the stress tensor
Research Faculty
Pioneering Green Chemistry and Nanomaterials for a Sustainable Future
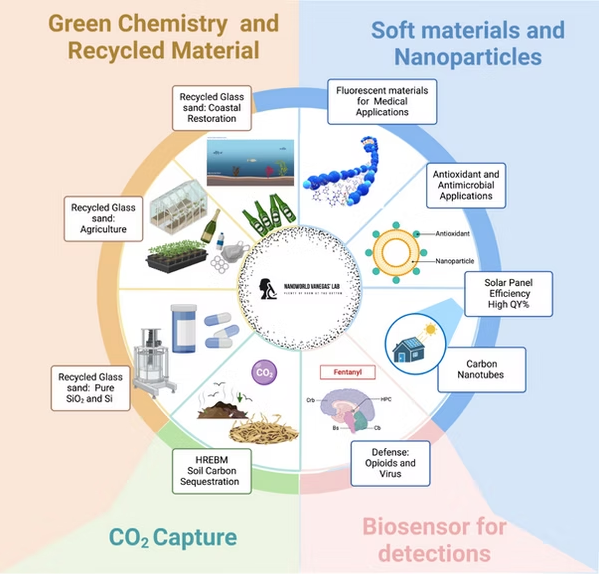 Biosensors for Detection, where advanced material manipulation techniques are employed to develop biosensors for the detection of bacteria, viruses, and compounds such as opioids, including fentanyl and its derivatives.
Biosensors for Detection, where advanced material manipulation techniques are employed to develop biosensors for the detection of bacteria, viruses, and compounds such as opioids, including fentanyl and its derivatives.
Green Chemistry and Recycled Materials is an internationally recognized line of work that focuses on utilizing recycled materials like glass and solar panels for applications in beach restoration, sustainable agriculture, and SiO₂ extraction for pharmaceutical uses.
Soft Materials and Nanoparticles, which involves the creation of plasmonic and fluorescent nanoparticles for medical and pharmaceutical applications.
A line of research, CO₂ Capture, explores innovative methods for capturing CO₂ using recycled glass and reactive high-energy ball milling (RHEBM).